Signal flow imaging using average connectome
Contents
1. Introduction
1.1. Scenario
VBMEG can make movies displaying signal flows between brain regions from source currents. Using source currents and an average anatomical connectivity, this tutorial introduces the procedure to make the signal flow movies. The source currents were estimated from MEG and EEG data during auditory, somatosensory, and visual tasks in M/EEG source imaging. The average connectivity was estimated from publicly available 18 subjects' diffusion MRIs. All the procedures will be conducted with scripts.
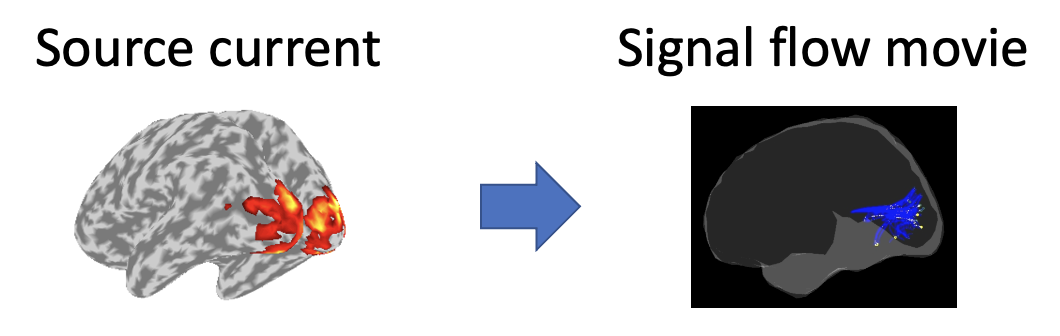
2. Starting tutorial
2.1. Setting environment
This tutorial was developed using MATLAB 2019a on Linux.
(1) Download and unzip tutorial.zip
(2) Copy and paste all the files to $your_dir
(3) Start MATLAB and change current directory to $your_dir/program
(4) Open make_all.m by typing
>> open make_all
2.2. Adding VBMEG to search path
Thereafter, we will sequentially execute the commands in make_all.m from the top.
We add VBMEG to search path.
>> path_of_VBMEG = '/home/cbi/takeda/analysis/toolbox/vbmeg_20220721';
>> addpath(path_of_VBMEG);
>> vbmeg
2.3. Set parameters
We set the parameters for modeling connectome dynamics, and set the file and directory names.
>> p = set_parameters;
By default, analyzed data and figures will be respectively saved in
- $your_dir/analyzed_data/s006,
- $your_dir/figure.
3. Signal flow imaging
We first load the average anatomical connectivity (3.1 Load anatomical connectivity). Then we estimate whole-brain dynamics models using the anatomical connectivity as a constraint (3.2. Modeling connectome dynamics). Finally we create movies displaying signal flows between region of interests (ROIs) (3.3. Creating movie).
3.1 Load anatomical connectivity
We load the average anatomical connectivity.
>> load_average_connectivity(p);
The following file will be saved.
- $your_dir/analyzed_data/s006/dmri/connectivity/connectivity.mat
3.2. Modeling connectome dynamics
From the source currents and the connectivity, we estimate a linear dynamics model (Connectome dynamics estimation).
Calculating ROI-current
We calculate ROI-current by extracting the principal component from the source currents within each ROI.
>> calculate_roi_current(p);
The following files will be saved.
- $your_dir/analyzed_data/s006/dynamics/roi_current/a.mat (Auditory task)
- $your_dir/analyzed_data/s006/dynamics/roi_current/s.mat (Somatosensory task)
- $your_dir/analyzed_data/s006/dynamics/roi_current/v.mat (Visual task)
Estimating dynamics model
From the ROI-current, we estimate a linear dynamics model using the L2-regularized least squares method. In this model, only the anatomically connected pairs of the ROIs have connectivity coefficients (Connectome dynamics estimation).
>> estimate_dynamics_model(p);
The following files will be saved.
- $your_dir/analyzed_data/s006/dynamics/model/a.lcd.mat (Auditory task)
- $your_dir/analyzed_data/s006/dynamics/model/s.lcd.mat (Somatosensory task)
- $your_dir/analyzed_data/s006/dynamics/model/v.lcd.mat (Visual task)
3.3. Creating movie
We create the movies displaying the signal flows between the ROIs (User manual).
Preparing input for creating movie
To create the movies, we need
- The ROI-current,
- The dynamics model,
- The place of the ROIs,
- Time information.
We gather these from separate files.
>> prepare_input_for_movie(p);
The following files will be saved.
- $your_dir/analyzed_data/dynamics/input_for_movie/a.mat (Auditory task)
- $your_dir/analyzed_data/dynamics/input_for_movie/s.mat (Somatosensory task)
- $your_dir/analyzed_data/dynamics/input_for_movie/v.mat (Visual task)
Loading average fiber shapes
To show signal flows along white matter fibers, we load their shapes between important ROIs. The fiber shapes were obtained from the 18 subjects' dMRIs.
>> load_average_fiber_shape(p);
The following files will be saved.
- $your_dir/analyzed_data/s006/dynamics/fiber/a.mat (Auditory task)
- $your_dir/analyzed_data/s006/dynamics/fiber/s.mat (Somatosensory task)
- $your_dir/analyzed_data/s006/dynamics/fiber/v.mat (Visual task)
Creating movie
We create the movies displaying the signal flows.
>> create_movie(p);
The following files will be saved.
- $your_dir/analyzed_data/s006/dynamics/movie/a/movie.avi (Auditory task)
- $your_dir/analyzed_data/s006/dynamics/movie/s/movie.avi (Somatosensory task)
- $your_dir/analyzed_data/s006/dynamics/movie/v/movie.avi (Visual task)
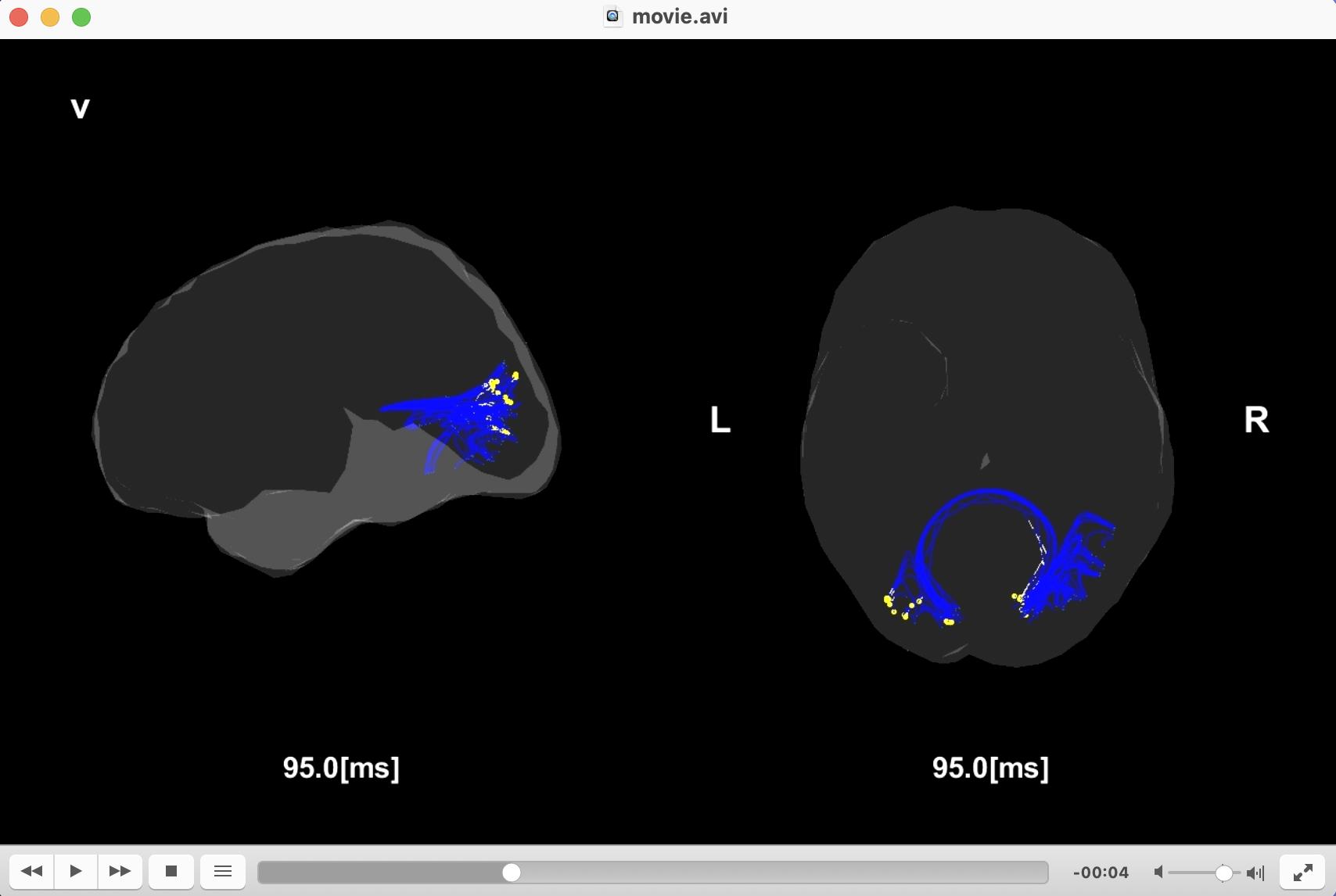
Congratulations! You have successfully achieved the goal of this tutorial.