ReleaseNotes
VBMEG v3.0.0-a-2 ReleaseNote
Nov 2023
- vb_make_fileinfo.m is added which funtion combine multiple MEG/EEG-files.
Channels are considered active or inactive, and the smallest available set of channels is active at merge time.
- AAL area/activity map is removed.
The standard brain (ICBM152) adopted by VBMEG and the defined image of the AAL area are different, it is difficult to resolve a discrepancy of a few millimeters, resulting in an imbalance in the number of cortical vertices in the left and right brain areas that is difficult to resolve.
VBMEG v3.0.0-a-1 ReleaseNote
May 2023
- Fixed error in calculating fibers for video when there are no anatomical connections between regions.
- Fixed error when loading extra dipole leadfield file(.basis.mat).
vb_load_basis(extra_basis_file);
- Fixed ignoring trial rejection flag(EEGinfo.ActiveTrial) when loading EEG-file(.eeg.mat).
- Fixed GUI plotting function that displays sensor and brain.(The standard brain was displayed instead of the individual brain).
- Supported the single-argument form of the following functions.
vb_job_leadfield_extra(extra_basis_parm);
vb_job_leadfield_extra_eeg(extra_basis_parm);
VBMEG v3.0.0-a-0 ReleaseNote
July 2022
What's New
- OPM data(QZFM) loading is implemented.
- The core module for DICOM conversion is replaced dcm2nii with dcm2niix.
- New cortical map - HCP MMP 1.0 (Glasser 2016) is supported.
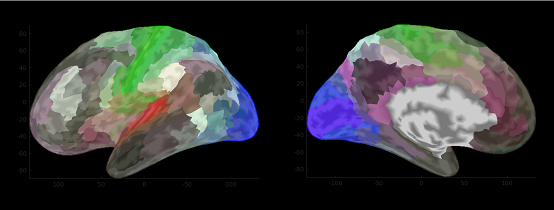
HCP_MMP1.area.mat - Diffusion MRI data processing using MRTrix3 is supported.
- VBMEG job function(vb_job_xxxx) with a single argument style is supported. The old style is still valid.
vb_job_xxxx(xxxx_parm); % single argument style(Newly supported)
vb_job_xxxx(proj_root, xxx_parm); % old style
VBMEG v2.2.0-a-1 ReleaseNote
Mar 2021
Important note: The standard brain model of v2.2.0-a-0 became freesurfer6.0-compatible. For this reason, the brain model of ver2.2 is not compatible with that of earlier version. More concretely, the number of vertex points is slightly smaller than that of ver2.0, 2.1.What's New
- bugfix
Dipole reduction ratio parameter doesn't work correctly. Duplicate cortex vertices are selected as activity points.
Patch file for v2.2.0-a-0 to v2.2.0-a-1 is here.
VBMEG v2.2.0-a-0 ReleaseNote
Feb 2020
Important note: The standard brain model of v2.2.0-a-0 became freesurfer6.0-compatible. For this reason, the brain model of ver2.2 is not compatible with that of earlier version. More concretely, the number of vertex points is slightly smaller than that of ver2.0, 2.1.What's New
- Compatibility : Supported MATLAB 2014b or later.
- Standard brain model has been changed from FreeSurfer 4.5 generated data to 6.0 generated data.
- Changes in area files created when importing a brain model.
- Deleted brodmann Cortical area file(brodmann.area.mat) because of less accuracy.
- Added new cortical area files by importing Freesurfer 6.0 annotation files.
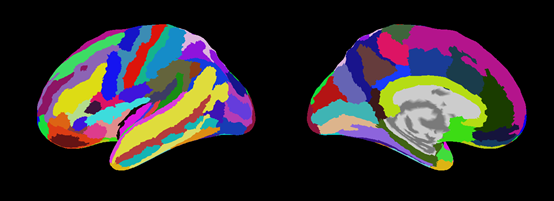
Destrieux.area.mat (aparc.a2009s.annot) 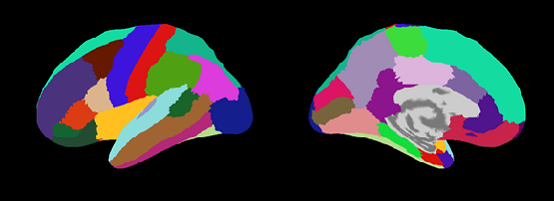
Desikan-Killiany.area.mat (aparc.annot) 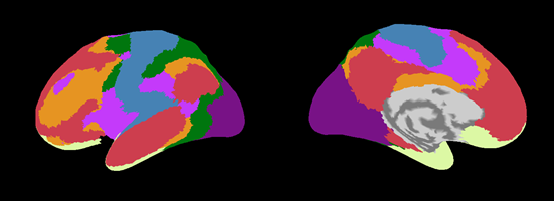
Yeo2011_7Networks.area.mat (Yeo2011_7Networks_N1000.annot) 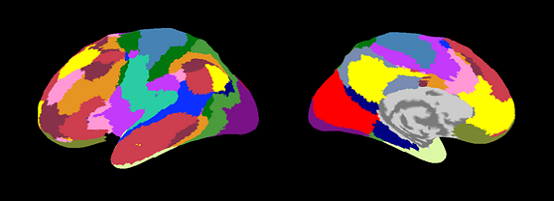
Yeo2011_17Networks.area.mat (Yeo2011_17Networks_N1000.annot)
- Changed surface reduction algorithm from MATLAB's reducepatch to iso2mesh(CGAL v3.5 Beta) to create surface model for BEM used in VBMEG.
NOTE
- The officially supported version of FreeSurfer has been changed to 6.0. Please use FreeSurfer 6.0 to process MRI structural images.
VBMEG v2.0-0-a-0 ReleaseNote
Dec 2017
This is VBMEG2.0, a major update to the VBMEG software, containing substantial functional enhancements over previous versions, as detailed in this document.
Main feature of VBMEG 2.0
- Connectome dynamics estimation
We have proposed a method to estimate current source dynamics under a constraint of whole-brain structural connectivity, called connectome (Fukushima et al. 2015, NeuroImage). The Fukushima's variational Bayesian method as well as two-step connectome dynamics estimation methods are implemented. These methods are applicable to investigate event-related or evoked brain activities.
- Visualization of estimated connectome dynamics as the dynamics movie
We propose a method to visualize complicated connectome dynamics as the dynamics movie. This method allows for visualizing connectome dynamics and current source activities in an unified way.
- MEG+EEG simultaneous current source imaging
To take advantage of complementary nature of MEG and EEG, MEG and EEG simultaneous current source imaging in the framework of VBMEG is implemented.
Detailed changes from version 1.0
Brain model
- From ver2.0, the MNI-aligned brain model is adopted to enhance group-level cortical source imaging analysis. Each vertex point in the new brain model is assigned with a fixed MNI coordinate of the standard brain (for example, vertex 1 = MNI coordinate [-69 -37 -2], vertex2 = [-67 -42 -2] and so on) while keeping cortical structure (normal direction) of each individual brain.
- Change specification of head model output file parameter (head_parm.head_file)
Only the specified head_parm.head_file is output, and no intermediate file is output.
- Default MRI file format is changed from Analyze(.hdr/.img) to NIfTI (.nii).
note : The MRI file used in VBMEG 2.0 is NIfTI format (.nii), the center coordinates are adjusted to the center of the image .This adjustment is done in the convert_dicom_nifti.m function which is DICOM format to NIfTI format. So NIfTI file used for analysis must always be created using this function ''.
- Add a function to execute FreeSurfer from MATLAB (vb_freesurfer_run.m).
- Change brain model import parameters
- spm_normalization_file for creating Broadmann and AAL area file became unnecessary.
- In order to remove corpus, specify FreeSurfer's label file (brain_parm.FS_left_label_file, .FS_right_label_file).
- Change format of Cortical model file (.brain.mat)
Added standard brain model to the Cortical model file(.brain.mat)
- Cortical area file(.area.mat)
The definition of 'Cortex' has been changed(VBMEG1.0: containing corpus, VBMEG2.0: No corpus) For this reason, the region where the current source is supposed is different between VBMEG 1.0 and 2.0.
fMRI data processing
- Automatic fMRI import
In VBMEG 1.0, it was necessary to operate SPM for each experiment condition and store it in the .spm.mat file, then import it one by one into the area/act file. This procedure was complicated and a cause of human errors. With VBMEG 2.0, it is now possible to automatically import all activity maps registered in the SPM result file (SPM.mat) into the area / act file. The imported activity map names become $conname_T (T value) and $conname_PS (Percent signal change). It is assumed that the SPM analysis is performed on the standard brain in default setting.
Current source imaging
- bayes_parm.Ta0_act and Ta0 are replaced with bayes_parm.prior_weight. In this new parameter, the prior confidence relative to the number of samples is specified with a real value ranging from 0 to 1 (0 : least confindence) .
- Added function vb_convert_current_unit.m for mutual conversion of the unit of time series data in the estimated current file(.curr.mat) (Amplitude [Am] <==> Density [Am / mm 2]).
Connectome dynamics
- Fukushima's dynamics estimation
- Add a function vb_job_vb_dynamics.m and related subfunctions.
- Add a function vb_job_current_dynamics.m which creates estimated current file (.curr.mat) from dynamics estimation result of Fukushima's method.
- Add Linear dynamics connectome estimation toolbox.
- Add functions for creating movie of the dynamics estimation result.
- Add diffusion MRI Processor toolbox in which structural connection strength and length matrices between brain regions are computed in cooperation with external tools (FreeSurfer, FSL, MRTrix).
Others
- project_mgr(GUI) is modified to be able to put stored parameter into M-script from the "Export as M-file".
Example exported M-script:
function test_job_brain
% original parameter(2017-02-09 11:48:40)
% Project root directory
proj_root = '/home/proj_root';
% Parameter
brain_parm.FS_left_file = '/home/FS_SUBJECT_DIR/Subject/bem/lh.smoothwm.asc';
brain_parm.FS_right_file = '/home/FS_SUBJECT_DIR/Subject/bem/rh.smoothwm.asc';
brain_parm.FS_left_infl_file = '/home/FS_SUBJECT_DIR/Subject/bem/lh.inflated.asc';
brain_parm.FS_right_infl_file = '/home/FS_SUBJECT_DIR/Subject/bem/rh.inflated.asc';
brain_parm.FS_left_curv_file = '/home/FS_SUBJECT_DIR/Subject/bem/lh.curv.asc';
brain_parm.FS_right_curv_file = '/home/FS_SUBJECT_DIR/Subject/bem/rh.curv.asc';
brain_parm.FS_left_sphere_file = '/home/FS_SUBJECT_DIR/Subject/bem/lh.sphere.reg.asc';
brain_parm.FS_right_sphere_file = '/home/FS_SUBJECT_DIR/Subject/bem/rh.sphere.reg.asc';
brain_parm.FS_sphere_key = 'sphere.reg';
brain_parm.FS_left_label_file = '/home/FS_SUBJECT_DIR/Subject/label/lh.cortex.label';
brain_parm.FS_right_label_file = '/home/FS_SUBJECT_DIR/Subject/label/rh.cortex.label';
brain_parm.registration_mode = 'FS';
brain_parm.analyze_file = '/home/subject/bolat/Four_quadrant_01/Subject/3D.hdr';
brain_parm.brain_file = './Subject/3D.brain.mat';
brain_parm.area_file = './Subject/3D.area.mat';
brain_parm.act_file = './Subject/3D.act.mat';
brain_parm.R_max = 0.018;
brain_parm.Nvertex = 10000;
brain_parm.reduce_ratio = 0.1;
brain_parm.N_step = 50;
brain_parm.display = 200;
brain_parm.brain_space = 'mni';
% Execute job function
vb_job_brain(proj_root, brain_parm);